Spectral Fit1
As of late September 2023, I do not have enough time to check this chapter in details. There might be a lot of bugs...
In order to perform spectral fit for spectra extracted from different annuli, we first have to create the spectra. In this chapter, we are going to learn how to do it, and the fitting will be performed in the next one.
To create annuli of suitable size for spectral fit, we need to define "suitable". Some people use number of counts, some use signal to noise(S/N) ratio. I suggest some readings first:
If you don't want to use cross arf, make sure the size of each annulus is > 30 arcsecs to limit flux redistribution to < 20% (Zhang 2007 Scaling relations and mass calibration of the X-ray luminous galaxy clusters at redshift ~0.2: XMM-Newton observations, Sec 2.3). In the same paper, the authors require a net counts of ~ 2000 in 2-7 keV for each spectrum.
Lovisari et al 2020 (X-Ray Scaling Relations for a Representative Sample of Planck-selected Clusters Observed with XMM-Newton) defined annulus size by S/N ratio and some other criteria. One purpose of deriving the spectrum of different annulus is to derive the temperature profile (for the derivation of hydrostatic mass). How many bins are enough? A good reference is Table 3 of this paper, column 11 NT. You see the number of bins can be as small as 1, and more than 30.
To derive the hydrostatic mass, you can use the backward method (based on a model like NFW profile) or forward method (no model assumed, I used this method for my work). In another paper by Lovisari et al 2020 (Comparing different mass estimators for a large subsample of the Planck-ESZ clusters), the less extrapolation of the temperature profile (i.e. last bin being closed to r500), the better the agreement between the hydrostatic masses derived using the forward and backward method. Again in Table 3 of the above mentioned paper, column 12 fT, you can check the fraction of r500 covered by the temperature profile. Also in the code I provide in this chapter, I provide the fraction of the mid point of the outermost annulus to the limiting radius you provide.
There are many others papers suitable. I will update this list later.
Here are what I use for this tutorial:
Source: MCXC J0106.8+0103
Observation ID: 0762870601
SAS Version: 19.0.0
Python version: 2 (yes, it is 2 instead of 3 this time. I think what you need to do is to change "import commands" to "import subprocess,shlex")
csh is used
There are quite a lot of stuff in this tutorial, I separate them into two files:
1. The main folder spectral_fit1: spectral_fit1.zip .
2. The folder products: products.zip . Just put this folder under the main one.
Only mos2 is used as an example.
You work in the main folder spectral_fit1, there are several subfolders:
1. src - the source code is found here.
2. InputFiles - the folder in which the good ccd lists for mos1(goodccdlist_mos1.dat) and mos2(goodccdlist_mos2.dat) are put. This is created in Anomalous CCD Check.
3.full_spectrum_mos2 - you created this folder previously in FOV spectrum and image. The images are in the energy range 0.4 - 2.3 keV. Now we need to create images in 0.7 - 10 keV and we need the products created before. I put them in this folder already.
4. products - the files created in this tutorial. The new files are put in the main folder if you create them successfully. To avoid the files being overwritten I create this new folder. Here again you find full_spectrum_mos2. I put the new files created in this chapter in products/full_spectrum_mos2. There is another folder mos2S002-0-30 in which you find the products created for the annulus [0 - 30] arcsec from the cluster centre.
5. command and log - these folders again contain old files needed to be used in this chapter. You find the same folders in products with new files inside.
If you have already finished the previous chapters and keep everything well. Then, you just need to copy the scripts in src to your current src folder.
Now let's begin with defining the size of an annulus by the number of counts, N, then
N = number_of_spectral_counts - number_of_background_counts
For S/N ratio (noise taken as error of Poisson statistics):
S/N = (number_of_spectral_counts - number_of_background_counts)/sqrt(number_of_spectral_counts)
Note: The spectrum includes both the FOV and background counts.
So in order the estimate the counts (or S/N) in a certain annulus, we need both the FOV and background spectra. The FOV spectrum takes seconds to produce but the background spectrum takes hours(you need to produce the corner spectra, images of each ccd chip...etc). To make things easier, I use the FOV and background images in the same energy range as the spectrum. For spectral anaylsis, I use 0.7 - 10keV, so now I extract images in the same range. Remember for the FOV spectrum and image, they are from the same source(i.e.mos2S002-clean.fits), so both the spectrum and image return you the same number of photons. But for the particle background spectrum and the particle background image, they are from different sources. However, in Table 2 of FOV Spectrum of Image, for the background spectrum and image in 0.7-10 keV, the mos ccds roughly have the same number of counts (pn is different but pn gives you enough counts anyway), so I think it is good to use the image data as an estimate.
Now we produce the images in the 0.7 - 10 keV using the script src/spectral_fit_script.py (remember to change your calibration path at the beginning of the script first):
python3 src/spectral_fit_script.py 1
"1" is step 1, which produces the following files in the folder full_spectrum_mos2:
mos2S002-obj-im-700-10000.fits (image of the FOV in sky coordinate)
mos2S002-obj-im-det-700-10000.fits (image of the FOV in detector coordinate)
mos2S002-im1-700-10000.fits (images of the ccd chip of the fwc files, only for good ccd chips)
mos2S002-im2-700-10000.fits
mos2S002-im3-700-10000.fits
mos2S002-im4-700-10000.fits
mos2S002-im5-700-10000.fits
mos2S002-im6-700-10000.fits
mos2S002-im7-700-10000.fits
mos2S002-exp-im-700-10000.fits (exposure image)
mos2S002-mask-im-700-10000.fits (an image indicating bad pixels and ccd gaps)
mos2S002-back-im-det-700-10000.fits (background image in detector coordinate)
mos2S002-back-im-sky-700-10000.fits (background image in sky coordinate)
comb-obj-im-700-10000.fits (mos2S002-obj-im-700-10000.fits*cheese)
comb-back-im-sky-700-10000.fits (mos2S002-back-im-sky-700-10000.fits*cheese)
comb-exp-im-700-10000.fits (mos2S002-exp-im-700-10000.fits*cheese corrected by scaling factor)
For pn, you also need the relevent oot images. Don't worry. The script has got it already.
We already produced the images in the soft band - 0.4 - 2.3 keV in FOV Spectrum and Image, and we wrote the commands in command/FOV_imgspe_mos2.csh . Now we are just doing the same thing. We first extract the relevant commands in command/FOV_imgspe_mos2.csh and rewrite them in full_spectrum_mos2/backgd_command_mos2.csh by replacing the old energy range with the new one. In the python script, full_spectrum_mos2/backgd_command_mos2.csh is run automatically and files are created in the same folder. In case of problems, run the commands in the csh script one by one to check where the problems are. Again note that for my csh script, the calibration path is mine, so you cannot run it directly.
Now we have produced the files in the energy range we want, it would be good to take a look at the surface brightness profile and check the size of the cluster. You can also take the r500 value in the MCXC catalogue as reference. The surface brightness profile also tells where you can choose the cosmic X-ray background. For me, I choose a region certain distance away from r500(say, 0.5 to 1 arcmin away). I will have more details in the next chapter Spectral Fit2 .
The problem comes here. We work in the range 0.7 - 10 keV, so we are supposed to check the profile in this range, right?
python3 src/spectral_fit_script.py 2a
("2a" plots the the surface brightness profile using the combined image, background and exposure map (i.e. comb-obj-im-700-10000-mos2.fits, comb-back-im-sky-700-10000-mos2.fits and comb-exp-im-700-10000-mos2.fits) in the folder full_spectrum_mos2 in 0.7 - 10 keV (surface brightness = (image - background)/exposure). In case you forget the meaning of "combined", check Fig. 10 of FOV spectrum and image)
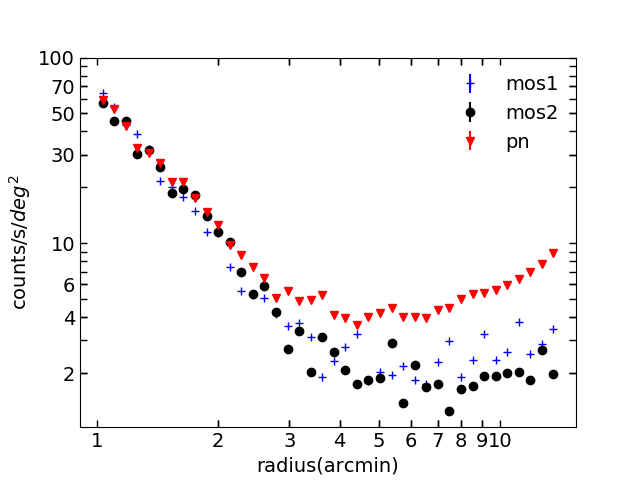 Fig.1 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.7 - 10.0 keV.
This cluster has a r500 of ~ 4.6 arcmins. The rising trend towards the end is due to the vignetting effect. Remeber the Sx = counts/exposure. Towards outer radius we have less and less counts, and the exposure gets smaller and smaller due to vignetting effect. The effect of smaller exposure outweighs the effect of less counts, resulting in a rising profile (I believe if you have a long enough exposure time, you will have sufficient counts to have a flat profile). You see this trend in most clusters.
I got off topic... the problem here is obviously pn has a higher cosmic X-ray background. So what about 0.4 - 2.3 keV?
python3 src/spectral_fit_script.py 2
("2" produces the same plot but in 0.4 - 2.3 keV)
Fig.1 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.7 - 10.0 keV.
This cluster has a r500 of ~ 4.6 arcmins. The rising trend towards the end is due to the vignetting effect. Remeber the Sx = counts/exposure. Towards outer radius we have less and less counts, and the exposure gets smaller and smaller due to vignetting effect. The effect of smaller exposure outweighs the effect of less counts, resulting in a rising profile (I believe if you have a long enough exposure time, you will have sufficient counts to have a flat profile). You see this trend in most clusters.
I got off topic... the problem here is obviously pn has a higher cosmic X-ray background. So what about 0.4 - 2.3 keV?
python3 src/spectral_fit_script.py 2
("2" produces the same plot but in 0.4 - 2.3 keV)
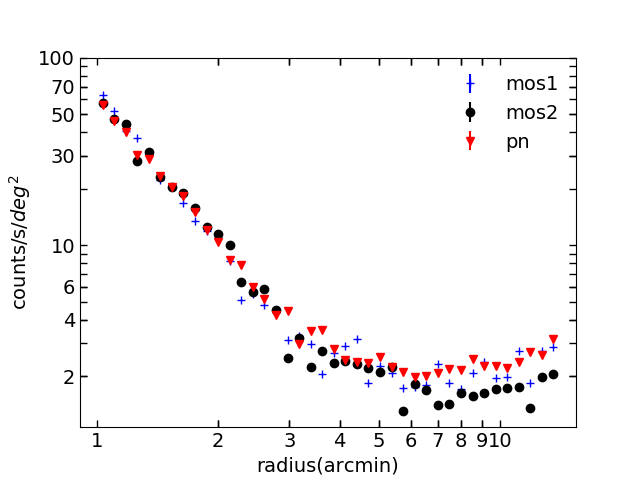 Fig.2 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.4 - 2.3 keV.
So you see here, for 0.4 - 2.3 keV, you have a pn profile which agress with mos, but not 0.7 - 10 keV. What does it mean? It is probably due to the particle background image. Let me remind you:
Sx = (counts - particle background)/exposure
which is
[(comb-obj-im-400-2300.fits) - (comb-back-im-sky-400-2300.fits)]/(comb-exp-im-700-400-2300.fits)
The difference in outer radius in the 0.7 - 10 keV plot is likely due to the particle background. Since this energy range shows noticeble difference I really wonder whether the particle background in this energy range correctly reflects the particle background ...Luckily for luminosity calculation, we only use the soft band. However, there is another problem, if the image correctly reflects the CXB background in all ccds, which mainly lies in low energy, what about the spectrum? In Table 2 of
FOV spectrum and image, I already showed that in the soft band, for the mos CCDs, the spectrum only have half of the background counts of the image (both are produced by mos_back) while pn has roughly the same number. If the background image in the soft band gives an accurante description of the background, then the spectrum wouldn't ...ok... I will check in the next chapter. What is affected here is the CXB value, an overestimation of which leads to underestimation of the APEC normalization of the cluster. If you use spectral data to do the luminosity, it is certainly affected.
By the way, 1) this is true for all sources, except for those with noticeable soft proton contamination. At this stage, the image still contains soft proton, which will be removed after spectra fit. I will come back to check the surface brightness profile in 0.7 - 10 keV again to see whether soft proton is successfully removed (OK! I found another way of checking for soft protons. This means. If soft proton outweighs particle background, pn and mos surface brightness profiles still overlap at the outskirt). 2) The problem comes from 7 - 10 keV, the high energy end (I checked the images in different energy bands but I am not showing the results here).
I need to go back to track ... from the above two plots (fig 1 and 2), the fortunate thing is the profiles for all ccds for any energy range becomes flat at more or less the same point, roughly 4 - 5 arcmins. Then we can decide where to choose the CXB. Here I use 4.6 arcmin (taken from the MCXC catalogue) as the cutoff radius for the cluster and 5.6 - 15 arcmin as the CXB background.
3. Now we have set our criteria, we can run the script (You will be asked at the prompt to input the limiting radius. Enter in arcsec. The value for this sample is 276.):
python3 src/spectral_fit_script.py 3
The criteria for the size of annulus is 1) > 1500 counts (I use mos2 as reference here) 2) The inner and outer radii should be at least 30 arcsecs apart(Zhang et al 2008). You can change the criteria by modifying the code (use of S/N instead of number of counts is ok).
The script returns you the following (the exact number may be slightly different):
source region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
0.00 - 30.00: || 6134.66 || 6659.19 || 14701.18
30.00 - 60.00: || 2785.77 || 2667.03 || 6127.48
60.00 - 110.00: || 1720.39 || 1665.47 || 3546.17
110.00 - 276.00: || 1640.75 || 1540.89 || 4329.92
S/N ratio
0.00 - 30.00: || 81.44 || 78.18 || 120.99
30.00 - 60.00: || 50.97 || 52.06 || 77.43
60.00 - 110.00: || 38.23 || 38.90 || 57.03
110.00 - 276.00: || 26.68 || 28.22 || 54.07
cxb region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
336.00 - 596.00: 1368.19 || 1504.17 || 9870.67
596.00 - 900.00: 1463.09 || 1537.47 || 13368.24
S/N ratio
336.00 - 596.00: 19.45 || 17.55 || 72.80
596.00 - 900.00: 16.89 || 15.40 || 83.53
source/bkg ratio
336.00 - 596.00: 0.38 || 0.26 || 1.25
596.00 - 900.00: 0.24 || 0.19 || 1.16
('last region(mid point) to limiting radius ratio: ', '0.70')
The result is written in spectral_region.dat
The final result is written in spectral_region.dat in the main folder. This file will be used when making the spectra. I provide the ratio of the mid-point of the last region to the limiting radius. As I said at the beginning, this ratio matters in the forward or backward method.
This is the annuli (in arcsecs) for the source region and CXB. We set the outermost annulus of the source to be 276 arcsecs. And the CXB region begins at 276 + 60 = 336 arcsecs. There are two regions found for the CXB since the criteria is fulfilled. You can modify them manually if you want only one region. Just change spectral_region.dat.
Now let's take a look at spectral_region.dat:
0.0 source
30.0 source
60.0 source
110.0 source
276.0 source
336.0 cxb
596.0 cxb
900.0 cxb
This file tells you what regions are for the source and what regions are for CXB.
Before we move on to produce the spectra, if you just want to check the number of counts in a particular radius, do step 4:
python3 bin/spectral_fit_script.py 4
Then you will be asked to input the inner and outer radius at the prompt, just input the values in arcsecs and you will get the number of counts and S/N ratio for the three ccds.
Finally, to produce the spectra, open 3 separate terminals and run the following 3 commands, one for each terminal:
source src/spectra-ann.csh mos1S001 path_to_calibration
source src/spectra-ann.csh mos2S002 path_to_calibration
source src/spectra-ann.csh pnS003 path_to_calibration
The script src/spectra-ann.csh does the following:
1) It reads spectral_region.dat and produces the spectrum one by one. A separate folder is created every time and an existing one is deleted. For example, the first spectrum in 0-30 arcsecs, a folder named mos2S002-0-30 is created and everything is produced there.
2) The files necessary for producing the spectrum is copied to the folder (i.e. mos2S002-clean.fits, mos2S002-bkg_region-sky.fits, mos2S002-bkg_region-det.fits and the oot files for pn)
3) Since we are not producing the whole FOV spectrum, we need to provide a region. The script writes this file as mos2-0-30.reg (in the folder mos2S002-0-30). Here is the content of mos2-0-30.reg:
&&((DETX,DETY) IN circle(232.60537,-57.590704,600))&&!((DETX,DETY) IN circle(232.60537,-57.590704,0))
The centre is in detector coordinate and is read from Centre.txt in the main folder. Here the centre is x = 232.6 and y = -57.59. 600 is the outer radius in pixels. Since one pixel = 0.05 arcsrc, 30 arcsecs translate to 600 pixels. The "0" is the inner radius. We work on the area "IN circle(232.60537,-57.590704,600))" but we excluse "IN circle(232.60537,-57.590704,0))".
4) The following commands are executed:
mos-spectra prefix=2S002 caldb=$path_to_calibration region=mos2-0-30.reg mask=1 elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2-spectra-0-30.log
mos_back prefix=2S002 caldb=$path_to_calibration elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2_back-0-30.log
To decide which ccd chip should be enabled, the file InputFiles/goodccdlist_mos2.dat is read. We set elow=0 ehigh=0 to produce the full energy range spectrum. If you only feel interested in a particular energy range, you can select it in the spectral fit when you use XSPEC.
The final products you want are: mos2S002-obj.pi, mos2S002-back.pi, mos2S002.rmf and mos2S002-back.arf. They are necessary for the spectral fit. The script renames them to mos2S002-obj-0-30.pi, mos2S002-back-0-30.pi, mos2S002-0-30.rmf and mos2S002-0-30.arf, respectively, and put them in the main folder.
The log files are written as log/mos2-spectra-0-30.log and log/mos2_back-0-30.log. If there is something wrong, check them. And the sub commands for mos-spectra are written in command/mos2-spectra-0-30.csh.
To make things faster, you can run all the spectra concurrently by opening multiple windows. Use the script src/spectra-single.csh:
source src/spectra-single.csh mos2S002 path_to_calibration 0 30
You just add the inner and outer radius in arcsecs and that particular spectrum would be produced.
Fig.2 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.4 - 2.3 keV.
So you see here, for 0.4 - 2.3 keV, you have a pn profile which agress with mos, but not 0.7 - 10 keV. What does it mean? It is probably due to the particle background image. Let me remind you:
Sx = (counts - particle background)/exposure
which is
[(comb-obj-im-400-2300.fits) - (comb-back-im-sky-400-2300.fits)]/(comb-exp-im-700-400-2300.fits)
The difference in outer radius in the 0.7 - 10 keV plot is likely due to the particle background. Since this energy range shows noticeble difference I really wonder whether the particle background in this energy range correctly reflects the particle background ...Luckily for luminosity calculation, we only use the soft band. However, there is another problem, if the image correctly reflects the CXB background in all ccds, which mainly lies in low energy, what about the spectrum? In Table 2 of
FOV spectrum and image, I already showed that in the soft band, for the mos CCDs, the spectrum only have half of the background counts of the image (both are produced by mos_back) while pn has roughly the same number. If the background image in the soft band gives an accurante description of the background, then the spectrum wouldn't ...ok... I will check in the next chapter. What is affected here is the CXB value, an overestimation of which leads to underestimation of the APEC normalization of the cluster. If you use spectral data to do the luminosity, it is certainly affected.
By the way, 1) this is true for all sources, except for those with noticeable soft proton contamination. At this stage, the image still contains soft proton, which will be removed after spectra fit. I will come back to check the surface brightness profile in 0.7 - 10 keV again to see whether soft proton is successfully removed (OK! I found another way of checking for soft protons. This means. If soft proton outweighs particle background, pn and mos surface brightness profiles still overlap at the outskirt). 2) The problem comes from 7 - 10 keV, the high energy end (I checked the images in different energy bands but I am not showing the results here).
I need to go back to track ... from the above two plots (fig 1 and 2), the fortunate thing is the profiles for all ccds for any energy range becomes flat at more or less the same point, roughly 4 - 5 arcmins. Then we can decide where to choose the CXB. Here I use 4.6 arcmin (taken from the MCXC catalogue) as the cutoff radius for the cluster and 5.6 - 15 arcmin as the CXB background.
3. Now we have set our criteria, we can run the script (You will be asked at the prompt to input the limiting radius. Enter in arcsec. The value for this sample is 276.):
python3 src/spectral_fit_script.py 3
The criteria for the size of annulus is 1) > 1500 counts (I use mos2 as reference here) 2) The inner and outer radii should be at least 30 arcsecs apart(Zhang et al 2008). You can change the criteria by modifying the code (use of S/N instead of number of counts is ok).
The script returns you the following (the exact number may be slightly different):
source region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
0.00 - 30.00: || 6134.66 || 6659.19 || 14701.18
30.00 - 60.00: || 2785.77 || 2667.03 || 6127.48
60.00 - 110.00: || 1720.39 || 1665.47 || 3546.17
110.00 - 276.00: || 1640.75 || 1540.89 || 4329.92
S/N ratio
0.00 - 30.00: || 81.44 || 78.18 || 120.99
30.00 - 60.00: || 50.97 || 52.06 || 77.43
60.00 - 110.00: || 38.23 || 38.90 || 57.03
110.00 - 276.00: || 26.68 || 28.22 || 54.07
cxb region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
336.00 - 596.00: 1368.19 || 1504.17 || 9870.67
596.00 - 900.00: 1463.09 || 1537.47 || 13368.24
S/N ratio
336.00 - 596.00: 19.45 || 17.55 || 72.80
596.00 - 900.00: 16.89 || 15.40 || 83.53
source/bkg ratio
336.00 - 596.00: 0.38 || 0.26 || 1.25
596.00 - 900.00: 0.24 || 0.19 || 1.16
('last region(mid point) to limiting radius ratio: ', '0.70')
The result is written in spectral_region.dat
The final result is written in spectral_region.dat in the main folder. This file will be used when making the spectra. I provide the ratio of the mid-point of the last region to the limiting radius. As I said at the beginning, this ratio matters in the forward or backward method.
This is the annuli (in arcsecs) for the source region and CXB. We set the outermost annulus of the source to be 276 arcsecs. And the CXB region begins at 276 + 60 = 336 arcsecs. There are two regions found for the CXB since the criteria is fulfilled. You can modify them manually if you want only one region. Just change spectral_region.dat.
Now let's take a look at spectral_region.dat:
0.0 source
30.0 source
60.0 source
110.0 source
276.0 source
336.0 cxb
596.0 cxb
900.0 cxb
This file tells you what regions are for the source and what regions are for CXB.
Before we move on to produce the spectra, if you just want to check the number of counts in a particular radius, do step 4:
python3 bin/spectral_fit_script.py 4
Then you will be asked to input the inner and outer radius at the prompt, just input the values in arcsecs and you will get the number of counts and S/N ratio for the three ccds.
Finally, to produce the spectra, open 3 separate terminals and run the following 3 commands, one for each terminal:
source src/spectra-ann.csh mos1S001 path_to_calibration
source src/spectra-ann.csh mos2S002 path_to_calibration
source src/spectra-ann.csh pnS003 path_to_calibration
The script src/spectra-ann.csh does the following:
1) It reads spectral_region.dat and produces the spectrum one by one. A separate folder is created every time and an existing one is deleted. For example, the first spectrum in 0-30 arcsecs, a folder named mos2S002-0-30 is created and everything is produced there.
2) The files necessary for producing the spectrum is copied to the folder (i.e. mos2S002-clean.fits, mos2S002-bkg_region-sky.fits, mos2S002-bkg_region-det.fits and the oot files for pn)
3) Since we are not producing the whole FOV spectrum, we need to provide a region. The script writes this file as mos2-0-30.reg (in the folder mos2S002-0-30). Here is the content of mos2-0-30.reg:
&&((DETX,DETY) IN circle(232.60537,-57.590704,600))&&!((DETX,DETY) IN circle(232.60537,-57.590704,0))
The centre is in detector coordinate and is read from Centre.txt in the main folder. Here the centre is x = 232.6 and y = -57.59. 600 is the outer radius in pixels. Since one pixel = 0.05 arcsrc, 30 arcsecs translate to 600 pixels. The "0" is the inner radius. We work on the area "IN circle(232.60537,-57.590704,600))" but we excluse "IN circle(232.60537,-57.590704,0))".
4) The following commands are executed:
mos-spectra prefix=2S002 caldb=$path_to_calibration region=mos2-0-30.reg mask=1 elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2-spectra-0-30.log
mos_back prefix=2S002 caldb=$path_to_calibration elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2_back-0-30.log
To decide which ccd chip should be enabled, the file InputFiles/goodccdlist_mos2.dat is read. We set elow=0 ehigh=0 to produce the full energy range spectrum. If you only feel interested in a particular energy range, you can select it in the spectral fit when you use XSPEC.
The final products you want are: mos2S002-obj.pi, mos2S002-back.pi, mos2S002.rmf and mos2S002-back.arf. They are necessary for the spectral fit. The script renames them to mos2S002-obj-0-30.pi, mos2S002-back-0-30.pi, mos2S002-0-30.rmf and mos2S002-0-30.arf, respectively, and put them in the main folder.
The log files are written as log/mos2-spectra-0-30.log and log/mos2_back-0-30.log. If there is something wrong, check them. And the sub commands for mos-spectra are written in command/mos2-spectra-0-30.csh.
To make things faster, you can run all the spectra concurrently by opening multiple windows. Use the script src/spectra-single.csh:
source src/spectra-single.csh mos2S002 path_to_calibration 0 30
You just add the inner and outer radius in arcsecs and that particular spectrum would be produced.
 Fig.1 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.7 - 10.0 keV.
This cluster has a r500 of ~ 4.6 arcmins. The rising trend towards the end is due to the vignetting effect. Remeber the Sx = counts/exposure. Towards outer radius we have less and less counts, and the exposure gets smaller and smaller due to vignetting effect. The effect of smaller exposure outweighs the effect of less counts, resulting in a rising profile (I believe if you have a long enough exposure time, you will have sufficient counts to have a flat profile). You see this trend in most clusters.
I got off topic... the problem here is obviously pn has a higher cosmic X-ray background. So what about 0.4 - 2.3 keV?
python3 src/spectral_fit_script.py 2
("2" produces the same plot but in 0.4 - 2.3 keV)
Fig.1 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.7 - 10.0 keV.
This cluster has a r500 of ~ 4.6 arcmins. The rising trend towards the end is due to the vignetting effect. Remeber the Sx = counts/exposure. Towards outer radius we have less and less counts, and the exposure gets smaller and smaller due to vignetting effect. The effect of smaller exposure outweighs the effect of less counts, resulting in a rising profile (I believe if you have a long enough exposure time, you will have sufficient counts to have a flat profile). You see this trend in most clusters.
I got off topic... the problem here is obviously pn has a higher cosmic X-ray background. So what about 0.4 - 2.3 keV?
python3 src/spectral_fit_script.py 2
("2" produces the same plot but in 0.4 - 2.3 keV)
 Fig.2 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.4 - 2.3 keV.
So you see here, for 0.4 - 2.3 keV, you have a pn profile which agress with mos, but not 0.7 - 10 keV. What does it mean? It is probably due to the particle background image. Let me remind you:
Sx = (counts - particle background)/exposure
which is
[(comb-obj-im-400-2300.fits) - (comb-back-im-sky-400-2300.fits)]/(comb-exp-im-700-400-2300.fits)
The difference in outer radius in the 0.7 - 10 keV plot is likely due to the particle background. Since this energy range shows noticeble difference I really wonder whether the particle background in this energy range correctly reflects the particle background ...Luckily for luminosity calculation, we only use the soft band. However, there is another problem, if the image correctly reflects the CXB background in all ccds, which mainly lies in low energy, what about the spectrum? In Table 2 of
FOV spectrum and image, I already showed that in the soft band, for the mos CCDs, the spectrum only have half of the background counts of the image (both are produced by mos_back) while pn has roughly the same number. If the background image in the soft band gives an accurante description of the background, then the spectrum wouldn't ...ok... I will check in the next chapter. What is affected here is the CXB value, an overestimation of which leads to underestimation of the APEC normalization of the cluster. If you use spectral data to do the luminosity, it is certainly affected.
By the way, 1) this is true for all sources, except for those with noticeable soft proton contamination. At this stage, the image still contains soft proton, which will be removed after spectra fit. I will come back to check the surface brightness profile in 0.7 - 10 keV again to see whether soft proton is successfully removed (OK! I found another way of checking for soft protons. This means. If soft proton outweighs particle background, pn and mos surface brightness profiles still overlap at the outskirt). 2) The problem comes from 7 - 10 keV, the high energy end (I checked the images in different energy bands but I am not showing the results here).
I need to go back to track ... from the above two plots (fig 1 and 2), the fortunate thing is the profiles for all ccds for any energy range becomes flat at more or less the same point, roughly 4 - 5 arcmins. Then we can decide where to choose the CXB. Here I use 4.6 arcmin (taken from the MCXC catalogue) as the cutoff radius for the cluster and 5.6 - 15 arcmin as the CXB background.
3. Now we have set our criteria, we can run the script (You will be asked at the prompt to input the limiting radius. Enter in arcsec. The value for this sample is 276.):
python3 src/spectral_fit_script.py 3
The criteria for the size of annulus is 1) > 1500 counts (I use mos2 as reference here) 2) The inner and outer radii should be at least 30 arcsecs apart(Zhang et al 2008). You can change the criteria by modifying the code (use of S/N instead of number of counts is ok).
The script returns you the following (the exact number may be slightly different):
source region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
0.00 - 30.00: || 6134.66 || 6659.19 || 14701.18
30.00 - 60.00: || 2785.77 || 2667.03 || 6127.48
60.00 - 110.00: || 1720.39 || 1665.47 || 3546.17
110.00 - 276.00: || 1640.75 || 1540.89 || 4329.92
S/N ratio
0.00 - 30.00: || 81.44 || 78.18 || 120.99
30.00 - 60.00: || 50.97 || 52.06 || 77.43
60.00 - 110.00: || 38.23 || 38.90 || 57.03
110.00 - 276.00: || 26.68 || 28.22 || 54.07
cxb region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
336.00 - 596.00: 1368.19 || 1504.17 || 9870.67
596.00 - 900.00: 1463.09 || 1537.47 || 13368.24
S/N ratio
336.00 - 596.00: 19.45 || 17.55 || 72.80
596.00 - 900.00: 16.89 || 15.40 || 83.53
source/bkg ratio
336.00 - 596.00: 0.38 || 0.26 || 1.25
596.00 - 900.00: 0.24 || 0.19 || 1.16
('last region(mid point) to limiting radius ratio: ', '0.70')
The result is written in spectral_region.dat
The final result is written in spectral_region.dat in the main folder. This file will be used when making the spectra. I provide the ratio of the mid-point of the last region to the limiting radius. As I said at the beginning, this ratio matters in the forward or backward method.
This is the annuli (in arcsecs) for the source region and CXB. We set the outermost annulus of the source to be 276 arcsecs. And the CXB region begins at 276 + 60 = 336 arcsecs. There are two regions found for the CXB since the criteria is fulfilled. You can modify them manually if you want only one region. Just change spectral_region.dat.
Now let's take a look at spectral_region.dat:
0.0 source
30.0 source
60.0 source
110.0 source
276.0 source
336.0 cxb
596.0 cxb
900.0 cxb
This file tells you what regions are for the source and what regions are for CXB.
Before we move on to produce the spectra, if you just want to check the number of counts in a particular radius, do step 4:
python3 bin/spectral_fit_script.py 4
Then you will be asked to input the inner and outer radius at the prompt, just input the values in arcsecs and you will get the number of counts and S/N ratio for the three ccds.
Finally, to produce the spectra, open 3 separate terminals and run the following 3 commands, one for each terminal:
source src/spectra-ann.csh mos1S001 path_to_calibration
source src/spectra-ann.csh mos2S002 path_to_calibration
source src/spectra-ann.csh pnS003 path_to_calibration
The script src/spectra-ann.csh does the following:
1) It reads spectral_region.dat and produces the spectrum one by one. A separate folder is created every time and an existing one is deleted. For example, the first spectrum in 0-30 arcsecs, a folder named mos2S002-0-30 is created and everything is produced there.
2) The files necessary for producing the spectrum is copied to the folder (i.e. mos2S002-clean.fits, mos2S002-bkg_region-sky.fits, mos2S002-bkg_region-det.fits and the oot files for pn)
3) Since we are not producing the whole FOV spectrum, we need to provide a region. The script writes this file as mos2-0-30.reg (in the folder mos2S002-0-30). Here is the content of mos2-0-30.reg:
&&((DETX,DETY) IN circle(232.60537,-57.590704,600))&&!((DETX,DETY) IN circle(232.60537,-57.590704,0))
The centre is in detector coordinate and is read from Centre.txt in the main folder. Here the centre is x = 232.6 and y = -57.59. 600 is the outer radius in pixels. Since one pixel = 0.05 arcsrc, 30 arcsecs translate to 600 pixels. The "0" is the inner radius. We work on the area "IN circle(232.60537,-57.590704,600))" but we excluse "IN circle(232.60537,-57.590704,0))".
4) The following commands are executed:
mos-spectra prefix=2S002 caldb=$path_to_calibration region=mos2-0-30.reg mask=1 elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2-spectra-0-30.log
mos_back prefix=2S002 caldb=$path_to_calibration elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2_back-0-30.log
To decide which ccd chip should be enabled, the file InputFiles/goodccdlist_mos2.dat is read. We set elow=0 ehigh=0 to produce the full energy range spectrum. If you only feel interested in a particular energy range, you can select it in the spectral fit when you use XSPEC.
The final products you want are: mos2S002-obj.pi, mos2S002-back.pi, mos2S002.rmf and mos2S002-back.arf. They are necessary for the spectral fit. The script renames them to mos2S002-obj-0-30.pi, mos2S002-back-0-30.pi, mos2S002-0-30.rmf and mos2S002-0-30.arf, respectively, and put them in the main folder.
The log files are written as log/mos2-spectra-0-30.log and log/mos2_back-0-30.log. If there is something wrong, check them. And the sub commands for mos-spectra are written in command/mos2-spectra-0-30.csh.
To make things faster, you can run all the spectra concurrently by opening multiple windows. Use the script src/spectra-single.csh:
source src/spectra-single.csh mos2S002 path_to_calibration 0 30
You just add the inner and outer radius in arcsecs and that particular spectrum would be produced.
Fig.2 Surface brightness profile of pnS003, mos1S001 and mos2S002 using combined images, background and exposure in 0.4 - 2.3 keV.
So you see here, for 0.4 - 2.3 keV, you have a pn profile which agress with mos, but not 0.7 - 10 keV. What does it mean? It is probably due to the particle background image. Let me remind you:
Sx = (counts - particle background)/exposure
which is
[(comb-obj-im-400-2300.fits) - (comb-back-im-sky-400-2300.fits)]/(comb-exp-im-700-400-2300.fits)
The difference in outer radius in the 0.7 - 10 keV plot is likely due to the particle background. Since this energy range shows noticeble difference I really wonder whether the particle background in this energy range correctly reflects the particle background ...Luckily for luminosity calculation, we only use the soft band. However, there is another problem, if the image correctly reflects the CXB background in all ccds, which mainly lies in low energy, what about the spectrum? In Table 2 of
FOV spectrum and image, I already showed that in the soft band, for the mos CCDs, the spectrum only have half of the background counts of the image (both are produced by mos_back) while pn has roughly the same number. If the background image in the soft band gives an accurante description of the background, then the spectrum wouldn't ...ok... I will check in the next chapter. What is affected here is the CXB value, an overestimation of which leads to underestimation of the APEC normalization of the cluster. If you use spectral data to do the luminosity, it is certainly affected.
By the way, 1) this is true for all sources, except for those with noticeable soft proton contamination. At this stage, the image still contains soft proton, which will be removed after spectra fit. I will come back to check the surface brightness profile in 0.7 - 10 keV again to see whether soft proton is successfully removed (OK! I found another way of checking for soft protons. This means. If soft proton outweighs particle background, pn and mos surface brightness profiles still overlap at the outskirt). 2) The problem comes from 7 - 10 keV, the high energy end (I checked the images in different energy bands but I am not showing the results here).
I need to go back to track ... from the above two plots (fig 1 and 2), the fortunate thing is the profiles for all ccds for any energy range becomes flat at more or less the same point, roughly 4 - 5 arcmins. Then we can decide where to choose the CXB. Here I use 4.6 arcmin (taken from the MCXC catalogue) as the cutoff radius for the cluster and 5.6 - 15 arcmin as the CXB background.
3. Now we have set our criteria, we can run the script (You will be asked at the prompt to input the limiting radius. Enter in arcsec. The value for this sample is 276.):
python3 src/spectral_fit_script.py 3
The criteria for the size of annulus is 1) > 1500 counts (I use mos2 as reference here) 2) The inner and outer radii should be at least 30 arcsecs apart(Zhang et al 2008). You can change the criteria by modifying the code (use of S/N instead of number of counts is ok).
The script returns you the following (the exact number may be slightly different):
source region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
0.00 - 30.00: || 6134.66 || 6659.19 || 14701.18
30.00 - 60.00: || 2785.77 || 2667.03 || 6127.48
60.00 - 110.00: || 1720.39 || 1665.47 || 3546.17
110.00 - 276.00: || 1640.75 || 1540.89 || 4329.92
S/N ratio
0.00 - 30.00: || 81.44 || 78.18 || 120.99
30.00 - 60.00: || 50.97 || 52.06 || 77.43
60.00 - 110.00: || 38.23 || 38.90 || 57.03
110.00 - 276.00: || 26.68 || 28.22 || 54.07
cxb region(arcsec) counts(bkg subtracted)
mos1 mos2 pn
336.00 - 596.00: 1368.19 || 1504.17 || 9870.67
596.00 - 900.00: 1463.09 || 1537.47 || 13368.24
S/N ratio
336.00 - 596.00: 19.45 || 17.55 || 72.80
596.00 - 900.00: 16.89 || 15.40 || 83.53
source/bkg ratio
336.00 - 596.00: 0.38 || 0.26 || 1.25
596.00 - 900.00: 0.24 || 0.19 || 1.16
('last region(mid point) to limiting radius ratio: ', '0.70')
The result is written in spectral_region.dat
The final result is written in spectral_region.dat in the main folder. This file will be used when making the spectra. I provide the ratio of the mid-point of the last region to the limiting radius. As I said at the beginning, this ratio matters in the forward or backward method.
This is the annuli (in arcsecs) for the source region and CXB. We set the outermost annulus of the source to be 276 arcsecs. And the CXB region begins at 276 + 60 = 336 arcsecs. There are two regions found for the CXB since the criteria is fulfilled. You can modify them manually if you want only one region. Just change spectral_region.dat.
Now let's take a look at spectral_region.dat:
0.0 source
30.0 source
60.0 source
110.0 source
276.0 source
336.0 cxb
596.0 cxb
900.0 cxb
This file tells you what regions are for the source and what regions are for CXB.
Before we move on to produce the spectra, if you just want to check the number of counts in a particular radius, do step 4:
python3 bin/spectral_fit_script.py 4
Then you will be asked to input the inner and outer radius at the prompt, just input the values in arcsecs and you will get the number of counts and S/N ratio for the three ccds.
Finally, to produce the spectra, open 3 separate terminals and run the following 3 commands, one for each terminal:
source src/spectra-ann.csh mos1S001 path_to_calibration
source src/spectra-ann.csh mos2S002 path_to_calibration
source src/spectra-ann.csh pnS003 path_to_calibration
The script src/spectra-ann.csh does the following:
1) It reads spectral_region.dat and produces the spectrum one by one. A separate folder is created every time and an existing one is deleted. For example, the first spectrum in 0-30 arcsecs, a folder named mos2S002-0-30 is created and everything is produced there.
2) The files necessary for producing the spectrum is copied to the folder (i.e. mos2S002-clean.fits, mos2S002-bkg_region-sky.fits, mos2S002-bkg_region-det.fits and the oot files for pn)
3) Since we are not producing the whole FOV spectrum, we need to provide a region. The script writes this file as mos2-0-30.reg (in the folder mos2S002-0-30). Here is the content of mos2-0-30.reg:
&&((DETX,DETY) IN circle(232.60537,-57.590704,600))&&!((DETX,DETY) IN circle(232.60537,-57.590704,0))
The centre is in detector coordinate and is read from Centre.txt in the main folder. Here the centre is x = 232.6 and y = -57.59. 600 is the outer radius in pixels. Since one pixel = 0.05 arcsrc, 30 arcsecs translate to 600 pixels. The "0" is the inner radius. We work on the area "IN circle(232.60537,-57.590704,600))" but we excluse "IN circle(232.60537,-57.590704,0))".
4) The following commands are executed:
mos-spectra prefix=2S002 caldb=$path_to_calibration region=mos2-0-30.reg mask=1 elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2-spectra-0-30.log
mos_back prefix=2S002 caldb=$path_to_calibration elow=0 ehigh=0 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& ../log/mos2_back-0-30.log
To decide which ccd chip should be enabled, the file InputFiles/goodccdlist_mos2.dat is read. We set elow=0 ehigh=0 to produce the full energy range spectrum. If you only feel interested in a particular energy range, you can select it in the spectral fit when you use XSPEC.
The final products you want are: mos2S002-obj.pi, mos2S002-back.pi, mos2S002.rmf and mos2S002-back.arf. They are necessary for the spectral fit. The script renames them to mos2S002-obj-0-30.pi, mos2S002-back-0-30.pi, mos2S002-0-30.rmf and mos2S002-0-30.arf, respectively, and put them in the main folder.
The log files are written as log/mos2-spectra-0-30.log and log/mos2_back-0-30.log. If there is something wrong, check them. And the sub commands for mos-spectra are written in command/mos2-spectra-0-30.csh.
To make things faster, you can run all the spectra concurrently by opening multiple windows. Use the script src/spectra-single.csh:
source src/spectra-single.csh mos2S002 path_to_calibration 0 30
You just add the inner and outer radius in arcsecs and that particular spectrum would be produced.