FOV spectrum and image
SAS Version: 19.0.0
(Remember to set your browser Tex-Encoding as unicode if some characters in this page is not probably displayed.)
Here are the subsections of this part:
Source spectrum and image production (
mos_spectrum
and
mos_back
)
Making background image in sky coordinate( rot-im-det-sky and esky2det )
Production of “Rate” image ( comb and bin_image )
Plotting radial profile using combined and non-combined exposure
Appendix - Error calculation of surface brightness profile.
Discussion:
1.Background counts of image vs spectrum
2.Should the particle background be divided by vignetting-corrected exposure?
FOV spectrum and image production
I put all the subcommands of mos-filter and pn-filter in command/FOV_imgspec_mos1.csh command/FOV_imgspec_mos2.csh command/FOV_imgspec_pn.csh I added comments to each subcommand. You can just extract any subcommand and run it. In the folder InputFiles , I already put the “good” ccd chips in a list called goodccdlist_mos1.dat and goodccdlist_mos2.dat (These are the results of Anomalous CCD Check ) , when you produce spectra of mos1 and mos2 , this file would be read automatically.
Now open three separate windows under the main folder, and run the following concurrently:
source bin/FOV_imgspec.csh mos1S001 400 2300 path_to_caldb
source bin/FOV_imgspec.csh mos2S002 400 2300 path_to_caldb
source bin/FOV_imgspec.csh pnS003 400 2300 path_to_caldb
You have to enter 4 arguments: 1: the full name* of the ccd, 2: lower limit of the energy (in eV) for the image you want to produce, 3: upper limit of the energy and 4: path to calibration data, i.e. where the fwc and other data is stored. Of course, for the energies and path to calibration data, you can modify the script and just put the values there.
*full name of the ccd is not necessarily mos1S001,mos2S002 or pnS003. Sometimes instead of “S”, it can be “U”, and it can be mos1S002, mos2S003 , pnS004 or something else.
For each of the ccd, a separate folder ( full_spectrum_mos2/mos1/pn ) is created and every process takes place in that folder. The file command.csh is updated all the time whenever a new process takes place. I rename it to FOV_imgspec_mos2(/mos1/pn).csh at the end of the whole process. The log files are in log/pn-spectra.log and mos-spectra_mos1(/mos2).log .We will use this file later. Now wait for a few hours (for my side mos1 takes the longest time ~10 hours, mos2 and pn ~ 6) for everything to finish to get all the stuff you need for this chapter, then you can go on with the rest. Note that I rename the files in the script.
Source spectrum and image production
To produce the spectrum and image, run the following, input the path of the calibration files in caldb :
mos-spectra prefix=2S002 caldb=path_to_caldb region=nonfile mask=1 elow=400 ehigh=2300 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1
pn-spectra prefix=S003 caldb=path_to_caldb region=nonfile mask=1 elow=1 ehigh=1 quad1=1 quad2=1 quad3=1 quad4=1
Now we want the whole FOV, so for the keyword region , just set nonfile . If mask=1 , mos-spectra will look for; mos2S002-bkg_region-det.fits and mos2S002-bkg_region-sky.fits (the same for mos1 and pn ) to make the spectrum and image with point sources masked. If these files are not present, spectrum and image would be produced with no mask. You can check log/mos-spectra-mos2.log to see whether the files are produced with them. It is written at the beginning. For mos1 and mos2, remember in the first chapter Filtering for Soft Protons , we find ccd5 of mos2 in anomaly. Set ccd5=0 would disable this ccd. For pn-spectra , just set all quad=1 .
A spectrum in the whole energy range is produced. Images in the energy range that you enter are produced. I choose 0.4-2.3 keV here because it is the soft band luminosity, a quantity I need to do my analysis. I will explain more about the choice of energy in the chapter (Luminosity Calculation) , where I will show you how to calculate luminosities.
Here are the brief steps for mos-spectra:
2.Production of spectrum of individual ccd chips using mos2S002-clean. fits(I skip mos1 and pn from now on)
3. Production of spectrum of individual ccd chip corners using mos2S002-clean.fits
5. Production of spectrum of individual ccd chip corners using FWC data
6. Production of image, exposure map and mask (not the cheese mask here) in the energy range of interest
7. Production of image of individual ccd chips using FWC data
For pn , the same is repeated with the oot file (i.e. pnS003-clean-oot.fits ).
Here are the details:
1.Production of spectrum in the field of interest, the relevant rmf and arf files using mos2S002-clean.fits.
To produce the whole FOV spectrum mos2S002-obj.pi :
evselect table=mos2S002-clean.fits:EVENTS withfilteredset=yes expression='(PATTERN<=12)&&(FLAG == 0)&&!((DETX,DETY) in BOX(-6560,13180,6590,6600,0))&®ion(mos2S002-bkg_region-det.fits)&&(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))' filtertype=expression keepfilteroutput=no updateexposure=yes filterexposure=yes withspectrumset=yes spectrumset=mos2S002-obj.pi energycolumn=PI spectralbinsize=5 withspecranges=yes specchannelmin=0 specchannelmax=11999
The expression is the most important. Pay attention to those involving coordinate selection. “A&&B” means overlapping area of A and B, and “A||B” means A+B (overlapping and non-overlapping area are both included), “!” means exclusion. So for the above:
(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))&&!((DETX,DETY) in BOX(-6560,13180,6590,6600,0))
means circle(-61,-228,17085)+ box(14.375,-16567.6,5552.62,795.625,0) (they together mean the exposed FOV) but excludes BOX(-6560,13180,6590,6600,0)
BOX(-6560,13180,6590,6600,0) corresponds to ccd5. You can extract an image using each of the circle or box in the expression to learn better. “ region(mos2S002-bkg_region-det.fits)” is the mask used. Finally,you extract a spectrum using this expression called mos2S002-obj.pi.
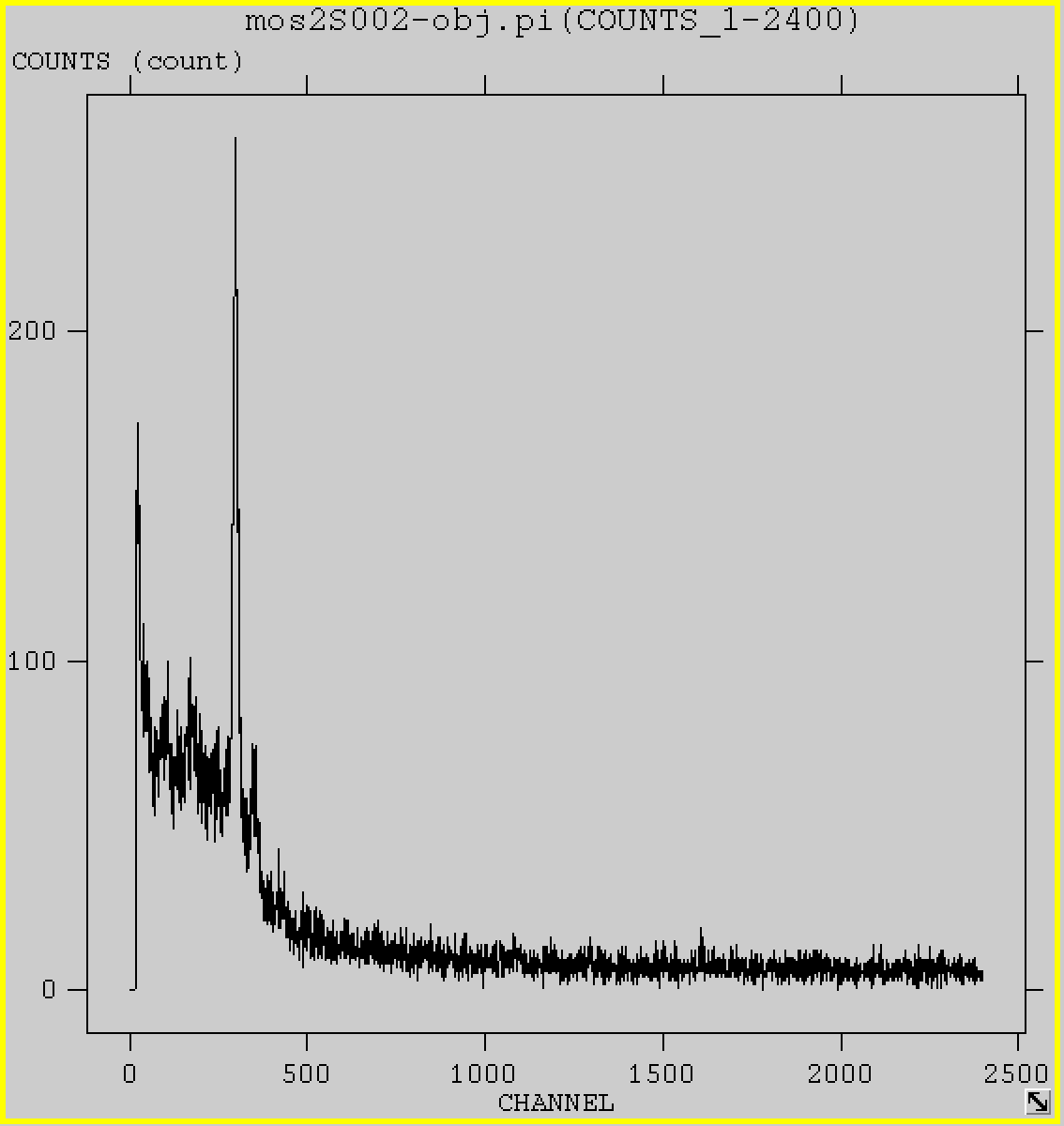
Now you open mos2S002-obj.pi to take a look at the header of the first extension SPECTRUM, look for SLCTEXPR , it is the selection expression that you enter. Explore some other keywords in the header yourself. Also check for the data of this extension, it only has two columns, counts and channel . Just make a plot directly with x=channel and y=counts, you plot the spectrum already!
fv mos2S002-obj.pi &
By the way, for image data, in case you forget what expression you use when you produce them, always check the header.
Here are the spectra of all ccd chips, including the anomalous ccd5. Here again you see at low energies, it has higher counts.
backscale spectrumset=mos2S002-obj.pi badpixlocation=mos2S002-clean.fits withbadpixcorr=yes
Now let’s check the value:
fkeyprint mos2S002-obj.pi+1 backscal
It is 714232892 square pixels. For the spectrum, 1 pixel = 0.05 arcsecs. So 714232892 would be 714232892*(0.05/60.)**2 = 496 square arcmins. You need this value in the spectral fit.
To produce the rmf file, which contains information about channel to energy conversion, in image called detmap.ds is first produced:
evselect table=mos2S002-clean.fits:EVENTS withfilteredset=yes filtertype=expression expression='(PATTERN<=12)&&(FLAG == 0)&&(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))&&!((DETX,DETY) in BOX(-6560,13180,6590,6600,0))&®ion(mos2S002-bkg_region-det.fits)' imagebinning='imageSize' imagedatatype='Int32' imageset=detmap.ds squarepixels=yes ignorelegallimits=no withxranges=yes withyranges=yes xcolumn='DETX' ximagesize=120 ximagemax=19500 ximagemin=-19499 ycolumn='DETY' yimagesize=120 yimagemax=19500 yimagemin=-19499 updateexposure=yes filterexposure=yes
rmfgen format=var rmfset=mos2S002.rmf spectrumset=mos2S002-obj.pi threshold=1.0e-6 detmaptype=dataset detmaparray=detmap.ds
You can now open mos2S002.rmf:
fv mos2S002.rmf &
See the second extension EBOUNDS. You see there are altogether 2400 channels and each channel corresponds to a particular energy range, with range = 5 keV. The mid energy of the first channel is 2.5keV, and the last 11.9975 keV.
and the exact detmap.ds is produced again for the arf file:
evselect table=mos2S002-clean.fits:EVENTS withfilteredset=yes filtertype=expression expression='(PATTERN<=12)&&(FLAG == 0)&&(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))&&!((DETX,DETY) in BOX(-6560,13180,6590,6600,0))&®ion(mos2S002-bkg_region-det.fits)' imagebinning='imageSize' imagedatatype='Int32' imageset=detmap.ds squarepixels=yes ignorelegallimits=no withxranges=yes withyranges=yes xcolumn='DETX' ximagesize=120 ximagemax=19500 ximagemin=-19499 ycolumn='DETY' yimagesize=120 yimagemax=19500 yimagemin=-19499 updateexposure=yes filterexposure=yes
then the arf:
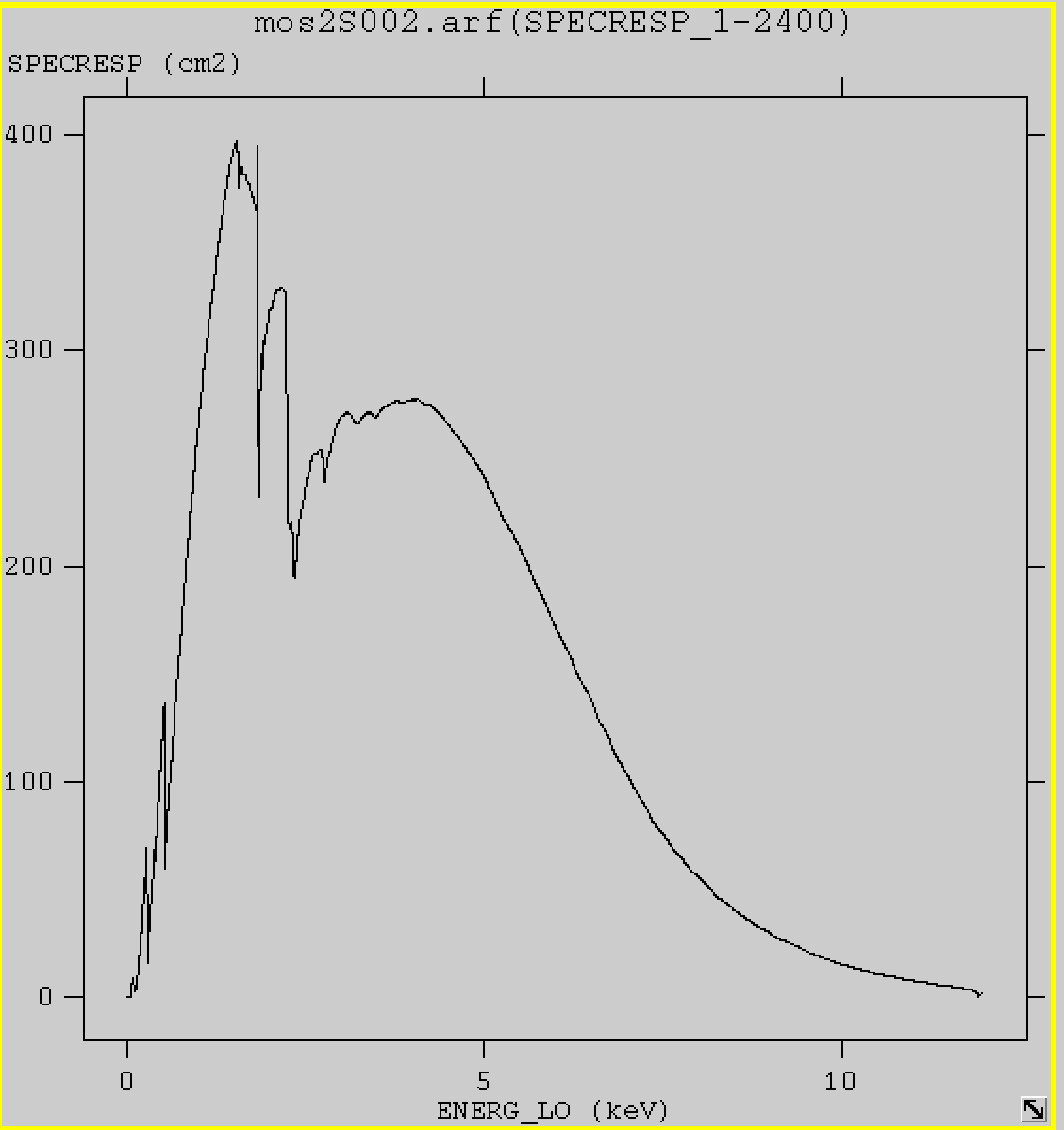
arfgen arfset=mos2S002.arf spectrumset=mos2S002-obj.pi withrmfset=yes rmfset=mos2S002.rmf extendedsource=yes modelee=no withbadpixcorr=no detmaptype=dataset detmaparray=detmap.ds badpixlocation=mos2S002-clean.fits modelootcorr=no
fv mos2S002.arf &
mos2S002.arf is basically the effective area curve. You see mos2 is most sensitive in the low energy range, ~ 1 – 5 keV. When you do spectral analysis, this has to be taken into account. This is the average effective area for the FOV. Later you will see there is difference in on-axis and off-axis positions.
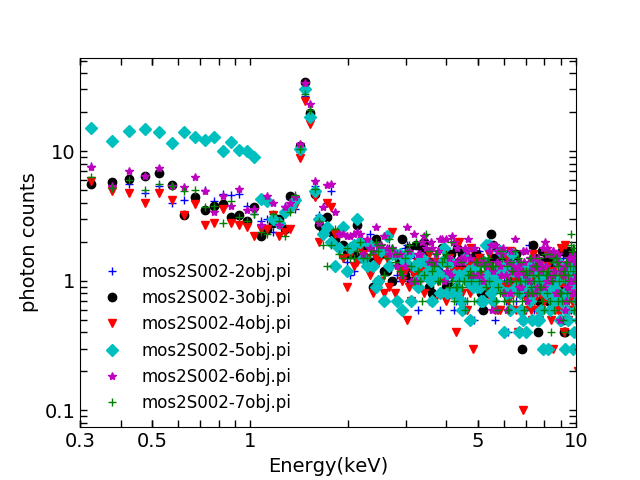
2.Production of spectrum of individual ccds using
mos2S002-clean.fits
. I use ccd2 as an example here:
evselect table=mos2S002-clean.fits:EVENTS withfilteredset=yes expression='(PATTERN<=12)&&(FLAG == 0)&&(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))&&((DETX,DETY) IN BOX(6580,-13530,6620,6640,0))&&!((DETX,DETY) in BOX(-6560,13180,6590,6600,0))&®ion(mos2S002-bkg_region-det.fits)' filtertype=expression keepfilteroutput=no updateexposure=yes filterexposure=yes withspectrumset=yes spectrumset=mos2S002-2obj.pi energycolumn=PI spectralbinsize=5 withspecranges=yes specchannelmin=0 specchannelmax=11999
Again for the next step you calculate the solid angle:
backscale spectrumset=mos2S002-2obj.pi badpixlocation=mos2S002-clean.fits withbadpixcorr=yes
4. Production of spectrum of individual ccd chip corners using
mos2S002-clean.fits
This is for the production of particle background (i.e.
mos2S002-back.pi
) in
mos_back
(to be carried out in the next part):
evselect table=mos2S002-corn.fits:EVENTS withfilteredset=yes expression='((DETX,DETY) IN BOX(6580,-13530,6620,6640,0))' filtertype=expression keepfilteroutput=no withspectrumset=yes spectrumset=mos2S002-2oc.pi energycolumn=PI spectralbinsize=5 withspecranges=yes specchannelmin=0 specchannelmax=11999
again the box is ccd2.
And then
backscale:
backscale spectrumset=mos2S002-2oc.pi useodfatt=no badpixlocation=mos2S002-clean.fits withbadpixcorr=yes ignoreoutoffov=no
Again this step is for mos_back . You can take a look at the FWC data called mos2-fwc.fits.gz. in the calibration folder. It is in the same format as mos2S002-clean. fits, but the data is taken with the filter wheel closed. It is a collection of different observations so there are a lot of counts:
evselect table=mos2-fwc.fits.gz:EVENTS withfilteredset=yes expression='(FLAG == 0)&&(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))&&((DETX,DETY) IN BOX(6580,-13530,6620,6640,0))&&!((DETX,DETY) in BOX(-6560,13180,6590,6600,0))&®ion(mos2S002-bkg_region-det.fits)' withspectrumset=yes keepfilteroutput=no updateexposure=yes filterexposure=yes spectrumset=mos2S002-2ff.pi energycolumn=PI spectralbinsize=5 withspecranges=yes specchannelmin=0 specchannelmax=11999
backscale spectrumset=mos2S002-2ff.pi badpixlocation=mos2S002-clean.fits withbadpixcorr=yes
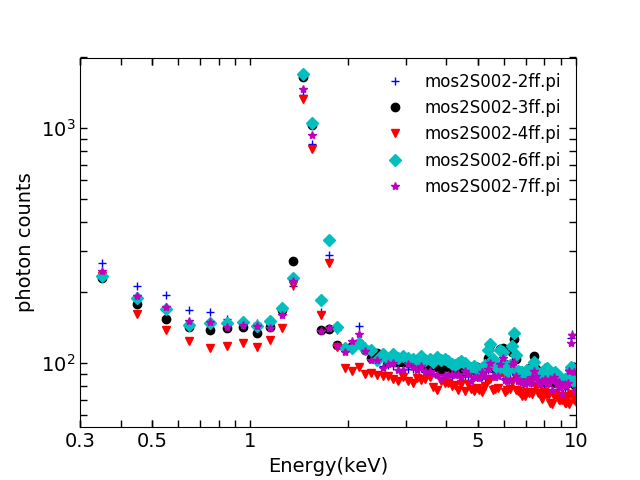
These spectra are called “ff”, e.g.
mos2S002-2ff.pi
. Here are all the ff spectra:
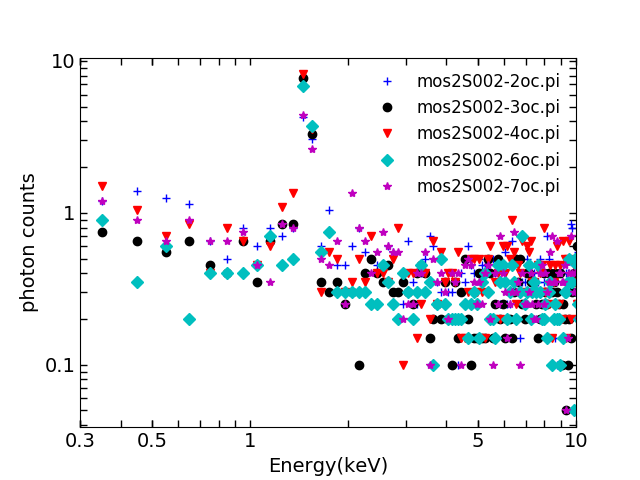
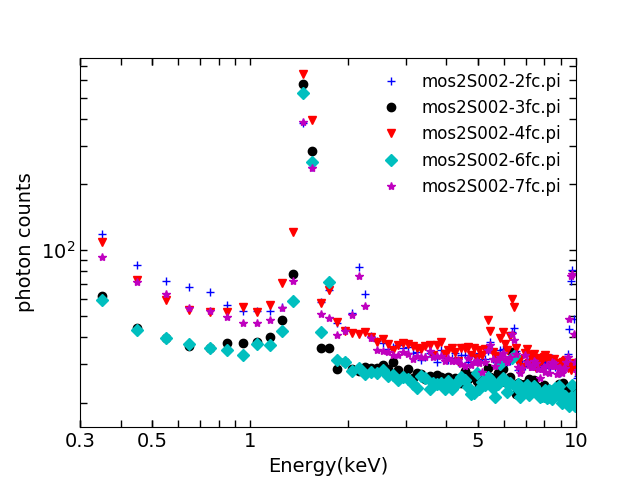
5. Production of spectrum of individual ccd corners using FWC data
These files are called “fc”,e.g.
mos2S002-2fc.pi
evselect table =mos2-fwc.fits.gz:EVENTS withfilteredset=yes expression='(PATTERN<=12)&&(FLAG == 65536)&&((DETX,DETY) IN BOX(6580,-13530,6620,6640,0))&&!(((DETX,DETY) IN CIRCLE(435,1006,17100))||((DETX,DETY) IN CIRCLE(-34,68,17700))||((DETX,DETY) IN BOX(-20,-17000,6500,500,0))||((DETX,DETY) IN BOX(5880,-20500,7500,1500,10))||((DETX,DETY) IN BOX(-5920,-20500,7500,1500,350))||((DETX,DETY) IN BOX(-20,-20000,5500,500,0)))' filtertype=expression keepfilteroutput=no withspectrumset=yes spectrumset=mos2S002-2fc.pi energycolumn=PI spectralbinsize=5 withspecranges=yes specchannelmin=0 specchannelmax=11999
backscale spectrumset=mos2S002-2fc.pi useodfatt=no badpixlocation=mos2S002-clean.fits withbadpixcorr=yes ignoreoutoffov=no
6. Production of image, exposure map and mask of the source in the energy range of interest
These are done using
mos2S002-clean.fits
which would be used to produce the “rate image”, i.e. surface brightness profile.
#image mos2S002-obj-im-400-2300.fits in [0.4-2.3]keV
evselect table=mos2S002-clean.fits:EVENTS withfilteredset=yes expression='(PATTERN<=12)&&(FLAG == 0) &&((CCDNR == 1)||(CCDNR == 2)||(CCDNR == 3)||(CCDNR == 4)||(CCDNR == 6)||(CCDNR == 7))&®ion(mos2S002-bkg_region-sky.fits)&&(PI in [400:2300])&&(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))' filtertype=expression imagebinning='imageSize' imagedatatype='Int32' imageset=mos2S002-obj-im-400-2300.fits squarepixels=yes withxranges=yes withyranges=yes xcolumn='X' ximagesize=900 ximagemax=48400 ximagemin=3401 ycolumn='Y' yimagesize=900 yimagemax=48400 yimagemin=3401 updateexposure=yes filterexposure=yes ignorelegallimits=yes
evselect table=mos2-fwc.fits.gz:EVENTS withfilteredset=yes expression='((DETX,DETY) IN BOX(6580,-13530,6620,6640,0))&&!((DETX,DETY) in BOX(-6560,13180,6590,6600,0))&®ion(mos2S002-bkg_region-det.fits)&&(FLAG == 0)&&(PI in [400:2300])&&(((DETX,DETY) IN circle(-61,-228,17085))||((DETX,DETY) IN box(14.375,-16567.6,5552.62,795.625,0)))' imagebinning='imageSize' imagedatatype='Int32' imageset=mos2S002-im2-400-2300.fits squarepixels=yes withxranges=yes withyranges=yes xcolumn='DETX' ximagesize=780 ximagemax=19500 ximagemin=-19499 ycolumn='DETY' yimagesize=780 yimagemax=19500 yimagemin=-19499 updateexposure=yes filterexposure=yes ignorelegallimits=yes
mos_back
mos_back prefix=2S002 caldb=path_to_calib_data diag=0 elow=400 ehigh=2300 ccd1=1 ccd2=1 ccd3=1 ccd4=1 ccd5=0 ccd6=1 ccd7=1 >& log/mos2_mos_back.log
After running
mos_back
, you see a file called
mos2S002-aug.qdp
. Let’s open it:
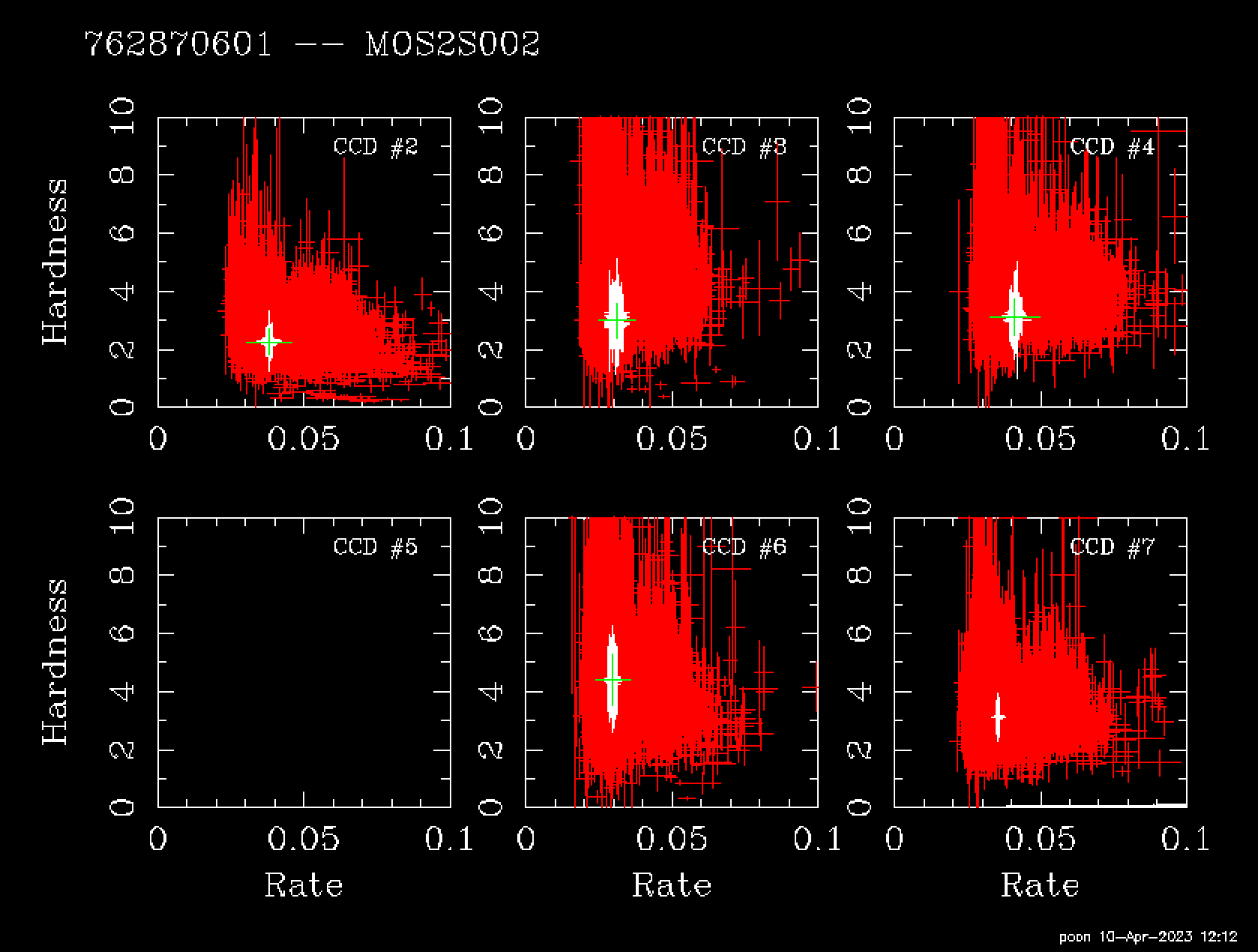
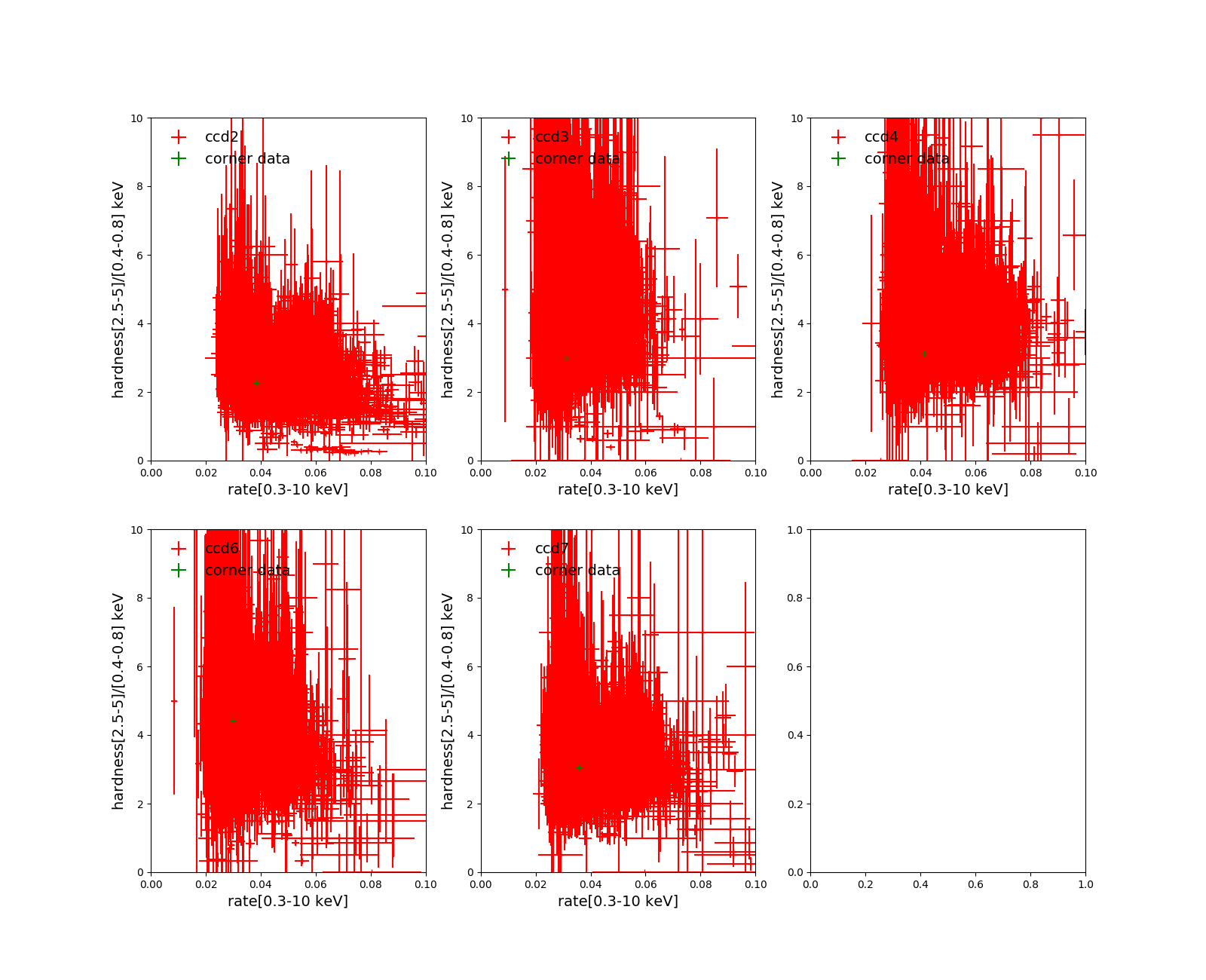
qpd mos2S002-aug.qdp
PGPLOT file/type:
/xw
Fig. 7 mos2S002-aug.qdp
The data in red is from the file mos2-qpb.fits.gz of the calibration data. Just open it and you see it has columns of “ HARD ” and “ RATE ”. The green cross is from our corner data with rate = (number of events in [0.3-10 keV])/(exposure) and hard =(number_of_events_in[2.5-5.0keV])/(number_of_events_in[0.40.8keV]). You can see these from the header of extension 0 of the file.
I don’t have the white + here as I don’t know the exact criteria how they are chosen, and this plot is somewhat different from mos2S002-aug.qdp probably due to different selection criteria used in both plots.
Finally,
scroll to almost the bottom of
log/mos2_mos_back.log
and I think this is where
mos2S002-back-im-det-400-2300.fits
is produced
.
I put the numbers in Table 1:
|
image
|
(A)Average non-zero value of FWC image
|
(B)Total number of counts in image
|
(C)FWC image scaling
|
C*S
|
|
mos2S002-im1-400-2300.fits
|
2.53404617
|
141478.531
|
1.23124784E-02
|
1741.95129
|
|
mos2S002-im2-400-2300.fits
|
2.59940314
|
101057.000
|
1.23224957E-02
|
1245.27441
|
|
mos2S002-im3-400-2300.fits
|
2.40861535
|
103355.812
|
1.21821780E-02
|
1259.09888
|
|
mos2S002-im4-400-2300.fits
|
2.49437261
|
89094.0000
|
1.16151366E-02
|
1034.83899
|
|
mos2S002-im6-400-2300.fits
|
2.47978711
|
110477.000
|
1.18284747E-02
|
1306.77441
|
|
mos2S002-im7-400-2300.fits
|
2.50920081
|
97893.5078
|
1.16683478E-02
|
1142.25549
|
|
Sum:
|
7730.19347
|
|||
(A)
The average non-zero value of FWC image = the total number of counts divided the number of non-zero pixels. Since this is from the calibration file;
mos2-fwc.fits.gz
, the value is source independent.
(B)
Total number of counts in image. Again this is from the calibration file
mos2-fwc.fits.gz
, the value is source independent.
(C)
FWC image scaling is related to the data exposure/fwc data exposure, i.e. exposure of [
mos2S002-1obj.pi/;
mos2S002-im1-400-2300.fits
]
Now let’s extract the exposure:
fkeyprint mos2S002-im1-400-2300.fits+0 EXPOSURE
fkeyprint mos2S002-1obj.pi+1 EXPOSURE
exposure for
mos2S002-im1-400-2300.fits:
1.91E+06s
exposure for mos2S002-1obj.pi: 2.70E+04s
So the ratio is 0.014, and it is basically the same for all ccds. Compare with the official value ~ 0.012, this is a little bit bigger. I guess the difference comes from the renormalization factor, as indicated in De Luca & Modelndi (2004) (Section 3.6), which "account for the different intensities of the NXB in the sky fields and in the closed observations due to the non-simultaneity of the measurement". And finally for the sum: 7730.19347, it is roughly the same as the total counts in mos2S002-back-im-det-400-2300.fits ,which is 7584.76758.
If you want to check the (A), (B), exposure ratio and total counts in mos2S002-back-im-det-400-2300.fits , in src/spectrum_script.py, do:
a.checkbkimg("mos2S002",[1,2,3,4,6,7])
Now we check the radial profile of
mos1S001-back-im-sky-400-2300.fits
mos2S002-back-im-sky-400-2300.fits
and
pnS003-back-im-sky-400-2300.fits
.
, with the centre in the central position of the image, i.e. xc=450, yc=450. Since in the table you see there is difference in the average non-zero value in the FWC image, the radial profile is not exactly that flat.
The ratio of (minimum bin/maximum bin) for mos1, mos2 and pn are ~ 87%,94% and 84%, respectively.
Making background image in sky coordinate
Now we have produced
mos2S002-back-im-det-400-2300.fits
,we want to produce the same image in sky coordinate, we do:
rot-im-det-sky prefix=2S002 mask=1 elow=400 ehigh=2300 mode=1 > mos2_rot_det_sky.log
esky2det datastyle=user ra=16.685125 dec=1.07061111111111 outunit=det withheader=no calinfostyle=set calinfoset=mos2S002-obj-im-400-2300.fits verbosity=0
The detector coordinate of the same position in X, Y are: 1232.412324, -1633.01633
You can read the ra and dec directly using ds9 or use ecoordconv :
ecoordconv imageset=mos2S002-obj-im-400-2300.fits x=450.91 y=450.91 coordtype=impix
You see on the screen you get the same ra, dec and detector coordinates:
ecoordconv:Region Centre:
Theta: Phi: 39.214786 2.6544148
X: Y: 25921 25921
DETX: DETY: 1232.4621 -1633.0042
IM_X: IM_Y: 450.91 450.91
RA: DEC: 16.685125 1.0706111
RAWX: RAWY: 356.4802 226.32278
CCD(s): 1 centred on CCD: 1
rot_det_sky mode=1 prefix=2S002 elow=400 ehigh=2300 detx=1232.412324 dety=-1633.01633 skyx=450.91 skyy=450.91 maskfile=1 clobber=1
And you are done!
comb caldb=$SAS_ESAS_CALDB withpartcontrol=1 withsoftcontrol=0 withswcxcontrol=0 elowlist=400 ehighlist=2300 mask=1 prefixlist="1S001" >&log/comb_mos1.log
mv comb-obj-im-400-2300.fits comb-obj-im-400-2300-mos1.fits
mv comb-back-im-sky-400-2300.fits comb-back-im-sky-400-2300-mos1.fits
mv comb-exp-im-400-2300.fits comb-exp-im-400-2300-mos1.fits
comb caldb=$SAS_ESAS_CALDB withpartcontrol=1 withsoftcontrol=0 withswcxcontrol=0 elowlist=400 ehighlist=2300 mask=1 prefixlist="2S002" >& log/comb_mos2.log
mv comb-obj-im-400-2300.fits comb-obj-im-400-2300-mos2.fits
mv comb-back-im-sky-400-2300.fits comb-back-im-sky-400-2300-mos2.fits
mv comb-exp-im-400-2300.fits comb-exp-im-400-2300-mos2.fits
comb caldb=$SAS_ESAS_CALDB withpartcontrol=1 withsoftcontrol=0 withswcxcontrol=0 elowlist=400 ehighlist=2300 mask=1 prefixlist="S003" >& log/comb_pn.log
mv comb-obj-im-400-2300.fits comb-obj-im-400-2300-pn.fits
mv comb-back-im-sky-400-2300.fits comb-back-im-sky-400-2300-pn.fits
mv comb-exp-im-400-2300.fits comb-exp-im-400-2300-pn.fits
bin_image thresholdmasking=0.02 detector=1 binning=1 prefix="2S002" elow=400 ehigh=2300 withpartcontrol=yes withsoftcontrol=no withswcxcontrol=no >& log/bin_image-mos2.log
The products are rate-400-2300.fits and sigma-400-2300.fits
This is just:
But to use them for radial profile, remember to multiply mos2S002-cheese.fits . In src/spectrum_script.py
a.rateimg("mos2S002-obj-im-400-2300.fits","mos2S002-back-im-sky-400-2300.fits","mos2S002-exp-im-400-2300.fits","mos2S002-cheese.fits","rate_mos2.fits
rate_mos2.fits
is the final product. The rate image
rate_mos2.fits
is corrected for
cheese
.
C is the conversion factor = 2073600 which is the unit conversion of pixel
2
to deg
2
, that means the unit of the rate image is counts/s/deg
2
. Remember 1 pixel = 2.5 arcsec = 2.5/3600. deg, so 1 deg
2
= (3600/2.5)
2
= 2073600.
For
pn,
the rate image is a little bit complicated as you have to deal with the
oot.
The rate image for
pn
is produced in this way:
where
pnS003-obj-im.fits = pnS003-obj-im-400-2300.fits
pnS003-back-im-sky.fits = pnS003-back-im-sky-400-2300.fits
pnS003-obj-im-oot.fits = pnS003-obj-im-oot-400-2300.fits
if (
(pnS003-obj-im-400-2300.fits - pnS003-obj-im-400-2300.fits*oot_scale_factor
) > 0:
else
sigma-400-2300.fits = 0
In src/spectrum_script.py :
a.rateimgpn("pnS003-obj-im-400-2300.fits","pnS003-obj-im-400-2300-oot.fits","pnS003-back-im-sky-400-2300.fits","pnS003-exp-im-400-2300.fits","pnS003-cheese.fits",0.023,"rate_pn.fits")
Of course, you don’t need rate-400-2300.fits and sigma-400-2300.fits for the real analyses. I just use the raw products in the code to allow more flexibility for units. Just remember to deal with the oot issue of pn . One thing to note:
comb-obj-im-400-2300-pn.fits = pnS003-obj-im-400-2300.fits - pnS003-obj-im-400-2300-oot.fits*oot_scale_factor
comb-obj-im-400-2300-pn.fits has already been corrected for oot, so I always use the combined image and particle background for surface brightness profile.
On the other hand, if you want to use the radial profile of the 3ccds with equal weighting, then you use comb-exp-im-400-2300.fits to produce the rate image. But before we make the radial profile, we first have to decide the centre. Here since the source is a relaxed cluster, we use the emission peak. In src/spectrum_script.py :
imgcomb()
emissionpeak( ) #parameters omitted
(For the emission peak and the emission-weighted centre, I will talk more in the Chapter Emission Peak vs Emission-weighted Centre )
The result is written in Centre.txt. It reads like this:
ra,dec = 16.706594 1.0560902
xc,yc(pix in python) = 420 430
detx,dety(mos1) = 160.70728 -402.16132
detx,dety(mos2) = 232.60537 -57.590704
detx,dety(pn) = -86.59832 -89.11581
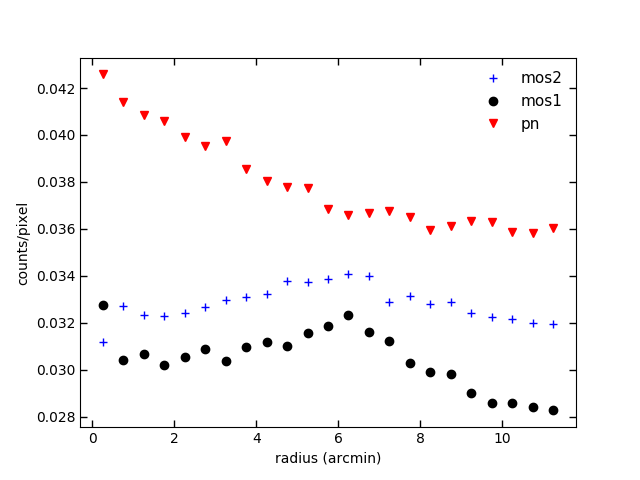
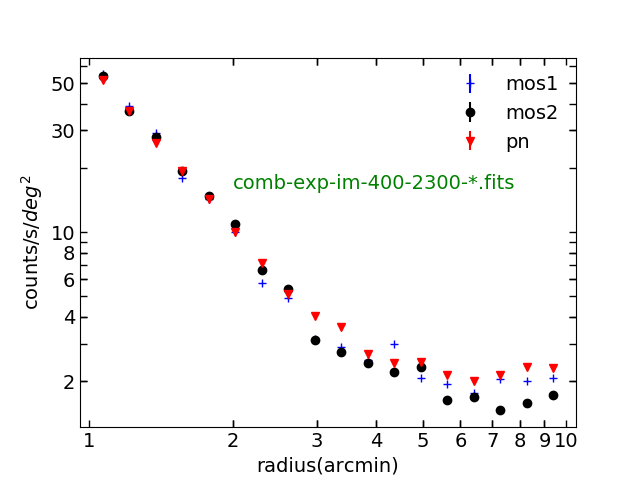
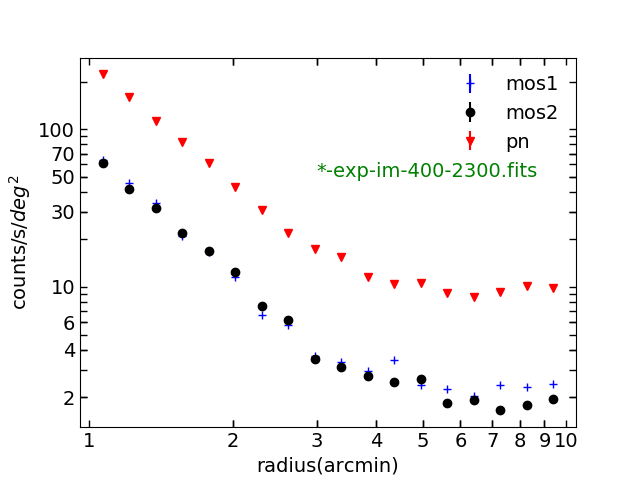
Plotting radial profile using combined and non-combined exposure
Now let’s make the radial profile using comb-exp-im-400-2300-mos2.fits and mos2S002 -exp-im-400-2300.fits and see the difference. In src/spectrum_script.py :
a.plotradial() #paramters omitted
Fig. 10. Surface brightness profile using the combined exposure, which is the uncombined one*correction_factor. mos1,mos2 and pn have the same weighting.
Fig. 11. Surface brightness profile using the uncombined exposure. mos1,mos2 and pn have different weightings.
By the way, you don’t see the errors bars in the surface brightness profiles. They are just too small. They are calculated using the standard error calculation. If you are unfamiliar with it, see the
Appendix
for “Error calculation of surface brightness profile”.
Now we have produced the background image
mos2S002-back-im-det-400-2300.fits
and background spectrum
mos2S002-back.pi
. The two are produced from different sources (the background image is from the fwc file and the spectrum is also related to the corner spectrum). I check whether they have the same counts in the same energy range. Since we need to first subtract the particle background from the source, correct estimation of the particle background directly affects CXB estimation, and hence the luminosity (You can derive the luminosity using either image data or spectral data).
I extracted the background images of
mos1
,
mos2
and
pn
in 1) 0.4-2.3 keV, 2) 0.7-10 keV and 3) 7.0-10.0 keV. The results are summarized in Table 2.
|
|
(Counts in spec/(counts in img) in [0.4-2.3] keV
|
(Counts in spec/(counts in img)
in [0.7-10.0] keV
|
(Counts in spec/(counts in img)
in [7.0-10.0] keV
|
|
mos1
|
~0.5
|
~1.0
|
~0.8
|
|
mos2
|
~0.5
|
~1.0
|
~0.8
|
|
pn
|
~1.0
|
~1.3
|
~2.9
|
Table 2: Summary of (counts in spectrum)/(counts in image) of different ccds in different energy range.
So for the energy range that a spectrum usually works on, [0.7-10.0] keV, the image and spectrum counts are quite consistent for all ccds, but for the range that the CXB lies in, [0.4-2.3] keV, the counts in image and spectrum are quite different for
mos1
and
mos2
, but not
pn
.
As for the range ([7.0-10.0] keV) that mainly proton flares are found,
mos1
and
mos2
find rather consistent count rates but not
pn
, which is ~3 times higher.
So how much does the particle background affect the r500 luminosity? The background rate is of the same magnitude as CXB (private study). Lowering the background rate would increase the CXB, but the increase and decrease wouldn’t be offset, as I will show in the Section Luminosity Calculation. I calculated the luminosities with the background image counts halved, and found for dim sources, the change in luminosity is ~20%. However, the current particle background of mos1, mos2 and pn give consistent luminosities across the three ccds, so I guess it is a good estimate.
However, the particle background is not affected by vignetting effect. As you can see in Table 1, column A, the counts/pixel is quite steady across different ccd chips, and ccd 1 does not have the highest value. I wonder whether the background should be divided by the vignetting-corrected exposure or vignetting-uncorrected exposure. I tested with vignetting-uncorrected exposure and find the change in luminosity minimal, a few percent at most.
P.S. If you want to produce vignetting-uncorrected exposure, when you do
eexpmap
, just set “
withvignetting=no”
Error calculation of surface brightness profile.
The error for the source is the square of source counts, which is the error for Poisson statistics. Then we apply the formular of error propagation:
So